If someone ever asks you what artificial intelligence has ever done for science, just show them AlphaFold. The program developed by Google’s AI group, known as DeepMind, has decoded the structure of almost all proteins in scientists’ catalogs, over 200 million of them. As the basic building blocks of life, proteins do most of the work in cells, from transmitting signals that regulate organs to protecting the body from bacteria and viruses. The ability to accurately predict the 3D structures of proteins from their amino-acid sequences is thus a huge boon to life sciences and medicine, and nothing short of revolutionary. This is a big deal because before AI scientists could only unravel the structure of a tiny fraction of these proteins.
Solving the protein folding problem
Proteins serve a wide range of purposes. Some are structural, others transport molecules, others still are receptors, and so on. Each of these functions is closely related to its specific shape, which is achieved through folding.
All proteins start off as a linear chain of basic units called amino acids. This primary 1D structure of amino acids contains the “recipe” that a protein uses to fold itself up. A protein will go through repeating stages of folding, adopting a wide range of configurations before reaching its final shape, which happens to be the most energetically favorable one.
However, predicting the 3D structure of a protein from its flat 1D sequence of amino acids is extremely challenging because the number of possible configurations can be staggering. Traditionally, structural biologists have determined protein structures through experimental means, using very expensive and time-consuming methods, such as X-ray crystallography or electron microscopy. Although accurate, this kind of research is very slow, hence we only knew about a few protein structures. But sifting through unfathomable amounts of possibilities for the human mind is exactly the kind of job an AI is best suited for.
DeepMind first revealed AlphaFold in 2020, and the scientific community was immediately blown away. Last year, in collaboration with the European Molecular Biology Laboratory (EMBL), DeepMind released a public database that included 98% of all human proteins, along with the protein structures for 20 other molecules.
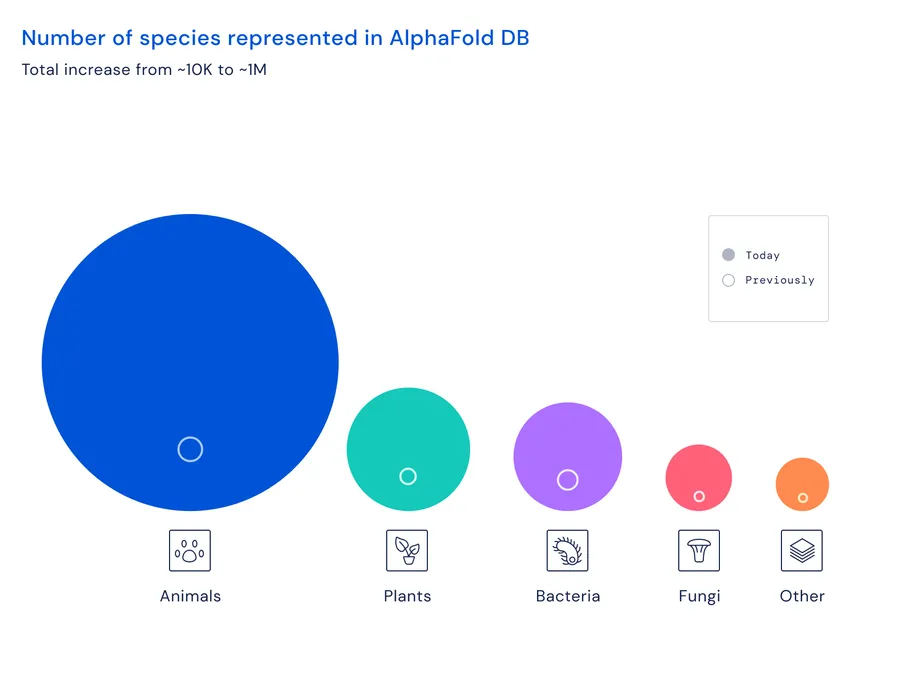
Now, the database has been expanded to cover all the proteins in almost every organism on Earth that has had its genome sequenced. That’s over 200 million structures.
“You can think of it as covering the entire protein universe,” Demis Hassabis, CEO of DeepMind, said during a press briefing. “We’re at the beginning of a new era now in digital biology.”
Less pipetting, more thinking
As genomic data is expected to swell like a tsunami each year, molecular biologists will have a field day with AlphaFold’s databases, empowering them to ask more advanced questions. For instance, armed with their 3D structures, scientists can now figure out the function of thousands of currently unsolved proteins in the human genome that may be linked to disease-causing gene variants that differ from person to person. They can also produce new drugs faster and respond to global threats like pandemics with greater zeal.
For instance, in early 2020, AlphaFold determined the structures of a handful of SARS-CoV-2 proteins that were determined experimentally. Imagine if a new dangerous pathogen is discovered tomorrow — AlphaFold would be able to quickly decipher its protein structure and rapidly arrive at possible avenues of attack in order to neutralize it.
Elsewhere, a research team led by Professor Matthew Higgins at the University of Oxford used AlphaFold’s predictions to unlock the structure of a key protein from a malaria parasite, allowing them to find the matching antibodies that can block the transmission of the parasite.
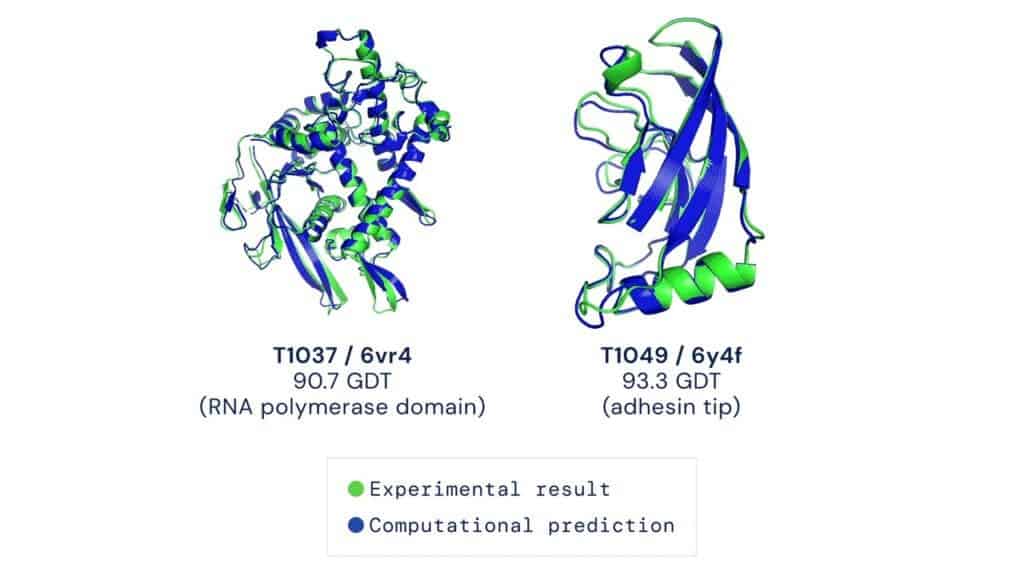
All of AlphaFold’s discovered protein structures, and even its source code, have been published for free. According to DeepMind, over 500,000 researchers from 190 countries have accessed the database so far, viewing two million structures.
However, all of this doesn’t mean the dawn of the experimental search for protein structures. AlphaFold is trained on datasets of protein structures that have been validated experimentally, and more such work is required to make the algorithm even more accurate. In fact, when dealing with highly challenging work, a hybrid approach combining technology and experimentation seems to work marvelously. Earlier this year, three research groups used AlphaFold to help them piece together one of the biggest jigsaw puzzles in biology, the human nuclear pore complex, which regulates the transport of macromolecules between the eukaryotic cell’s nucleus and cytoplasm and is composed of over 1,000 protein subunits.
“Its delicate structure was finally revealed by using existing experimental methods to reveal its outline and AlphaFold predictions to complete and interpret any areas that were unclear. This powerful combination is now becoming routine in labs, unlocking new science and showing how experimental and computational techniques can work together,” the DeepMind team wrote in a blog post.






