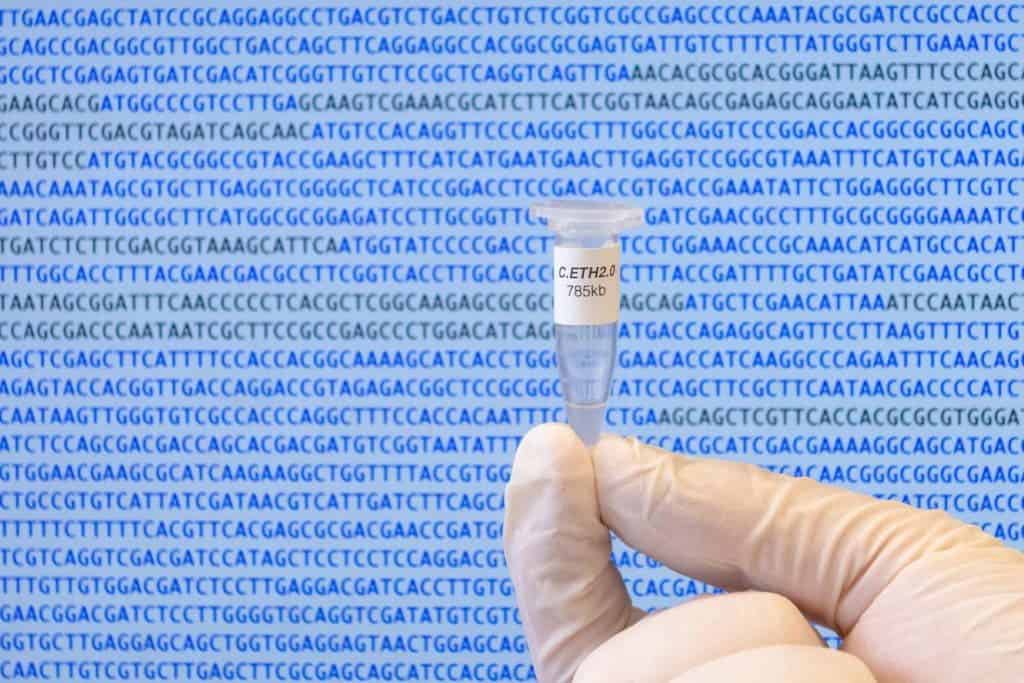
Researchers at ETH Zurich have demonstrated a new method for producing genomes that is cheaper and faster than ever before. The authors produced the first fully computer-generated genome — the Caulobacter ethensis-2.0 for which a corresponding organism does not yet exist. However, the genome was physically produced and inserted into an existing organism with similar genetic material.
Computers and synthetic life
In 2010, researchers at the J. Craig Venter Institute reported a landmark advancement in synthetic biology: the first bacterial DNA engineered from scratch. This was the culmination of more than a decade of hard work and a $40 million investment. But while the bacterial genome made by Craig Venter was an exact copy of a natural genome, researchers at ETH Zurich radically altered the genome of a model organism called Caulobacter crescentus. What’s more, their research spanned a time frame of a year and cost les than half a million dollars, which shows that synthetic biology is on the brink of a revolution.
Caulobacter crescentus is a harmless freshwater bacterium whose genome has been extensively studied. Previous research showed that out of the bacterium’s 4,000 genes only about 680 were crucial to the survival of the organism in laboratory conditions. Beat Christen, Professor of Experimental Systems Biology at ETH Zurich, and his brother, Matthias Christen, a chemist at ETH Zurich, used this minimum set of crucial genes as a starting point.
The researchers designed a computer algorithm that scanned this minimally viable natural genome and computed the ideal DNA sequence for the synthesis and construction of the genome. The algorithm replaced a sixth of all the 800,000 DNA letters found in the minimal genome.
“Through our algorithm, we have completely rewritten our genome into a new sequence of DNA letters that no longer resembles the original sequence. However, the biological function at the protein level remains the same,” says Beat Christen.
The hard part was only just beginning. Next, the researchers had to produce a DNA molecule which contained the artificial bacterial genome, and this had to be done step by step. The researchers synthesized 236 separate genome segments, which they then had to delicately piece together.
“The synthesis of these segments is not always easy,” explains Matthias Christen. “DNA molecules not only possess the ability to stick to other DNA molecules, but depending on the sequence, they can also twist themselves into loops and knots, which can hamper the production process or render manufacturing impossible,” explains Matthias Christen.
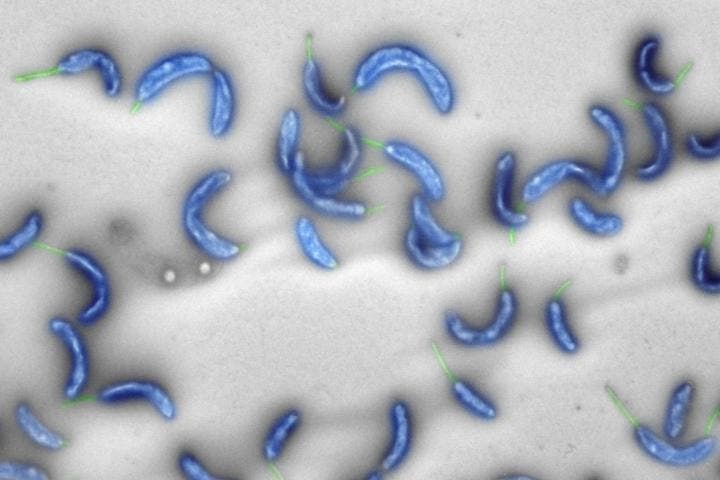
As an experiment, the researchers produced strains of bacteria in the lab that contained the naturally occurring Caulobacter genome as well as segments of the new artificial genome. By switching off certain natural genes in the bacteria, the researchers were able to test the functions of the artificial genes introduced earlier. The rewritten genome was designed by an algorithm which could only parse information that was understood at the time of the DNA sequence. Naturally, there are also DNA sequences that have yet to be understood by scientists, and this can be lost in the process of creating the new code.
“Our method is a litmus test to see whether we biologists have correctly understood genetics, and it allows us to highlight possible gaps in our knowledge,” explains Beat Christen.
These experiments showed that only 580 of the 680 artificial genes were functional, showing that the algorithm needs tweaking before researchers can hope to achieve a truly functional genome in version 3.0.
Even though this version isn’t perfect, the new study demonstrates how modern technology can streamline artificial DNA synthesis. And who knows: in the future scientists might finally create synthetic organisms that would serve a wide array of biotech applications. For instance, custom-made bacteria could be used to produce active molecule for drugs or DNA vaccines.
“We believe that it will also soon be possible to produce functional bacterial cells with such a genome,” says Beat Christen.
“As promising as the research results and possible applications may be, they demand a profound discussion in society about the purposes for which this technology can be used and, at the same time, about how abuses can be prevented,” he added. It is still not clear when the first bacterium with an artificial genome will be produced – but it is now clear that it can and will be developed. “We must use the time we have for intensive discussions among scientists, and also in society as a whole. We stand ready to contribute to that discussion, with all of the know-how we possess.”
The findings were reported in the Proceedings of the National Academy of Sciences.



