
The recent renaissance of the field of artificial intelligence after the slump of the 1980s (the so-called ‘AI winter’) has proven absolutely revolutionary. But perhaps there is no greater disruption caused by machine learning and other AI systems than in biology. It was just recently that DeepMind, a Google subsidiary tasked with developing cutting-edge AI programs, revealed that its AlphaFold system had decoded the structure of virtually all proteins known to science — over 200 million of them.
Now, a new AI tool is pushing the boundaries of what’s possible even further by allowing scientists to design original proteins from scratch unlike anything seen in nature. This novel tool, known as ProteinMPNN, was recently described by researchers at the University of Washington in a pair of studies published today in Science. Its authors are confident that ProteinMPNN and other similar tools that are bound to surface in the near future will open up a new realm of possibility and subsequent applications. These include entirely novel proteins that are designed from the ground up to meet a certain goal, whether it’s enzymes that digest plastic or new drugs that target some of today’s most challenging and intractable diseases.
“Proteins are fundamental across biology, but we know that all the proteins found in every plant, animal, and microbe make up far less than one percent of what is possible. With these new software tools, researchers should be able to find solutions to long-standing challenges in medicine, energy, and technology,” said senior author David Baker, professor of biochemistry at the University of Washington School of Medicine and recipient of a 2021 Breakthrough Prize in Life Sciences.
From discovering protein function to forging new proteins
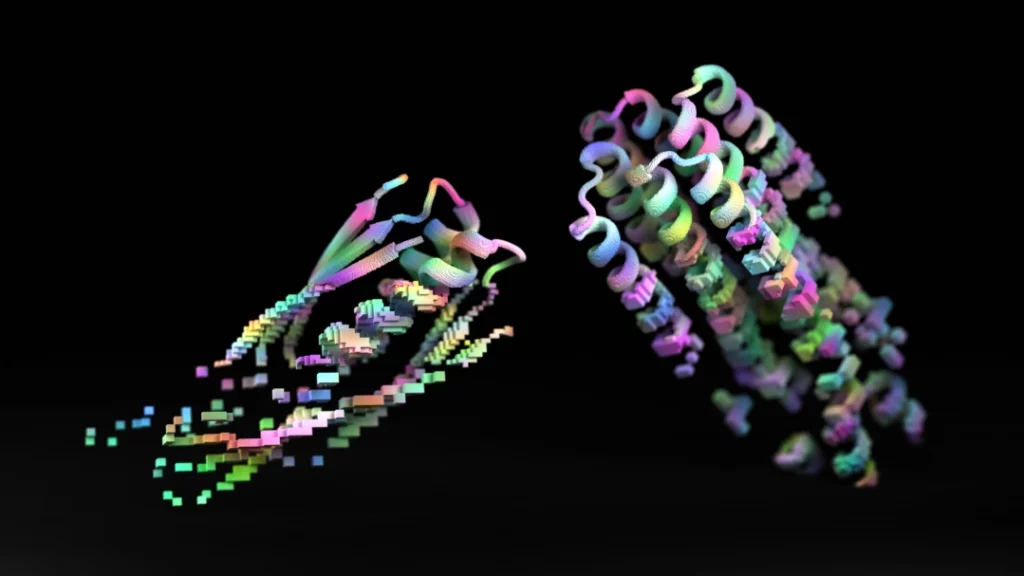
The role of proteins in supporting life and nature at large cannot be understated. Some are structural, others transport molecules, others still are receptors, and so on. Each of these functions is closely related to its specific shape, which is achieved through folding.
All proteins start off as a linear chain of basic units called amino acids. This primary 2D structure of amino acids contains the “recipe” that a protein uses to fold itself up. A protein will go through repeating stages of folding, adopting a wide range of configurations before reaching its final shape, which happens to be the most energetically favorable one.
While AlphaFold can predict the shapes of existing proteins, thereby deducing their function, ProteinMPNN tackles the same problem but from the opposite angle. Rather than reverse engineering the role of a protein from nature, the new tool can help scientists engineer entirely novel proteins from the ground up. They can, for instance, dream up a certain function or purpose for a protein, and then have the AI come up with the corresponding structure whose molecular components and shape are conducive to the desired function. It’s then a matter of simply synthesizing these proteins in the lab.
ProteinMPNN can achieve all of these truly remarkable things using two powerful AIs developed at the University of Washington. The first, dubbed “hallucination”, allows scientists to search among potentially useful protein sequences based on simple prompts — sort of like the now famous DALL-E generative AI tool that generates fantastic images from a text prompt. The second AI, known as “inpainting”, can be seen as an autocomplete feature like the kind you see when you type a question into Google — only it’s for proteins. When used in synergy, the two methods can enable scientists to discover entirely new proteins that fit a desired function.
In order to validate the different protein shapes generated by the two AIs, the researchers turned to the tried and tested AlphaFold to see whether the amino acid sequences were indeed likely to fold in the desired shape.
“ProteinMPNN is to protein design what AlphaFold was to protein structure prediction,” Baker said in a statement.
Proteins initially designed using ProteinMPNN were then assembled in the lab. Among them were nanoscale rings, each with a diameter a billion times smaller than a poppy seed, that could be fitted inside custom nanomachines.
“This is the very beginning of machine learning in protein design. In the coming months, we will be working to improve these tools to create even more dynamic and functional proteins,” said Baker.
ProteinMPNN is available for free and on GitHub.


