Researchers at the University of Texas at Austin’s Texas Advanced Computing Center (TACC) are simulating the SARS-COV-2 coronavirus envelope atom-by-atom to help us devise new treatments.
The effort is being led by Rommie Amaro, a professor of chemistry and biochemistry at the University of California, San Diego. The plan is to complete an all-atom model of the virus’s exterior envelope, the part that is involved in cell infection and drug interaction.
Judge a virus by its cover
“If we have a good model for what the outside of the particle looks like and how it behaves, we’re going to get a good view of the different components that are involved in molecular recognition,” says Amaro.
Since the model is anticipated to contain a whopping 200 million atoms, and the interactions of each with one another have to be calculated, the team called on Frontera, the #5 top supercomputer in the world and #1 academic supercomputer (according to November 2019 rankings of the Top500 organization) at the TACC, and some creative coding to help ease the burden.
“We’re trying to combine data at different resolutions into one cohesive model that can be simulated on leadership-class facilities like Frontera,” Amaro said. “We basically start with the individual components, where their structures have been resolved at atomic or near atomic resolution. We carefully get each of these components up and running and into a state where they are stable.”
“Then we can introduce them into the bigger envelope simulations with neighboring molecules.”
The team is very happy to have access to Frontera, as few other computers could tackle the sheer weight of calculations needed to run these models. The team says their current objective is to keep improving the performance of the simulation to allow for more tests to be run quickly.
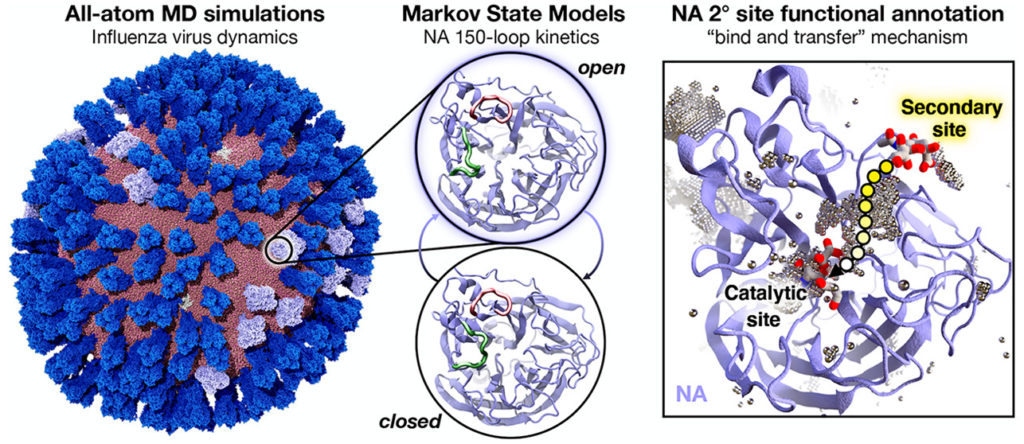
The model is based on Amaro’s previous work with an all-atom simulation of the influenza virus envelope which was published in February.
“These simulations will give us new insights into the different parts of the coronavirus that are required for infectivity,” says Amaro. “The information that we get from these simulations is multifaceted and multidimensional and will be of use for scientists on the front lines immediately and also in the longer term.”
“Hopefully the public will understand that there’s many different components and facets of science to push forward to understand this virus. These simulations on Frontera are just one of those components.”
The paper “Mesoscale All-Atom Influenza Virus Simulations Suggest New Substrate Binding Mechanism” has been published in the journal ACS Central Science.



