Within every biological body, there are thousands of proteins, each twisted and folded into a unique shape. The formation of these shapes is crucial to their function, and researchers have struggled for decades to predict exactly how this folding will take place.
Now, AlphaFold (the same AI that mastered the games of chess and Go) seems to have solved this problem, essentially paving the way for a new revolution in biology. But not everyone’s buying it.
What the big deal is
Proteins are essential to life, supporting practically all its functions, a DeepMind blog post reads. The Google-owned lab British artificial intelligence (AI) research became famous in recent years as their algorithm became the best chess player on the planet, and even surpassed humans in Go — a feat once thought impossible. After toying with a few more games, the DeepMind team set its eyes on a real-life task: protein folding.
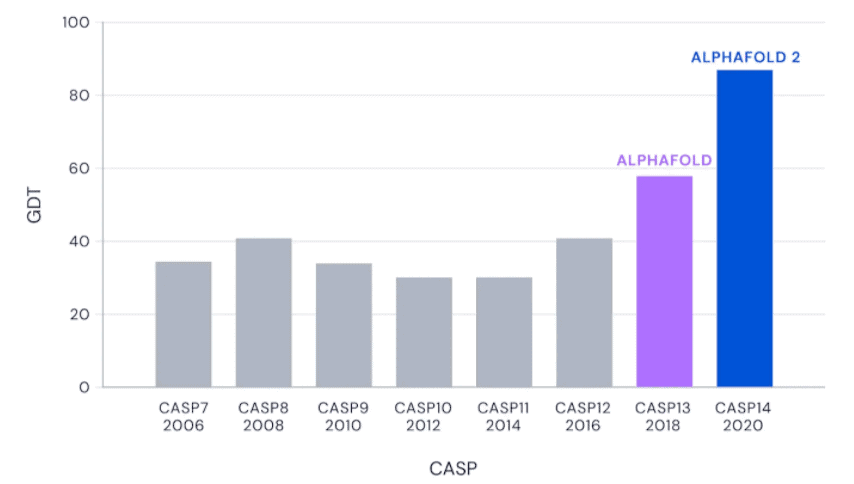
In 2018, the team announced that AlphaFold 2 (the second version of the protein folding algorithm) has become quite good at predicting the 3D shapes of proteins, surpassing all other algorithms. Now, two years later, the algorithm seems to have been perfected even more.
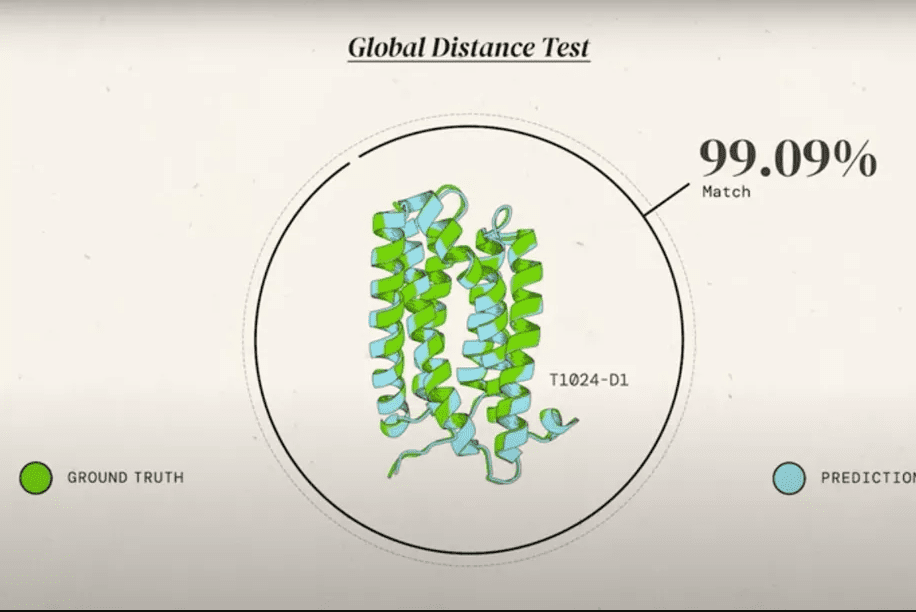
In a global competition called Critical Assessment of protein Structure Prediction, or CASP, AlphaFold 2 and other systems are given the amino acid strings for proteins and asked to predict their shape. The competition organizers already know the actual shape of the protein, but of course, they keep it secret. Then, the prediction is compared to real-world results. DeepMind CEO Demis Hassabis calls this the “Olympics of protein folding” in a video.
AlphaFold nailed it. Not all its predictions were spot on, but all were very close — it was the closest thing to perfection ever seen since CASP kicked off.
“AlphaFold’s astonishingly accurate models have allowed us to solve a protein structure we were stuck on for close to a decade,” Andrei Lupas, Director of the Max Planck Institute for Developmental Biology and a CASP assessor, said in the DeepMind blog.
CASP uses the “Global Distance Test (GDT)” metric, assessing accuracy from 0 to 100. AlphaFold 2 achieved a median score of 92.4 across all targets, which translates to an average error of approximately 1.6 Angstroms, or about the width of an atom.
It’s not perfect. Even one Angstrom can be to big of an error and render the protein useless, or even worse. But the fact that it’s so close suggests that a solution is in sight. The problem has seemed unsolvable for so long that researchers were understandably excited.
“We have been stuck on this one problem – how do proteins fold up – for nearly 50 years. To see DeepMind produce a solution for this, having worked personally on this problem for so long and after so many stops and starts, wondering if we’d ever get there, is a very special moment.”
Why protein folding is so important
It can take years for a research team to identify the shape of individual proteins — and these shapes are crucial for biological research and drug development.
A protein’s shape is closely linked to the way it works. If you understand its shape, you also have a pretty good idea of how it works.
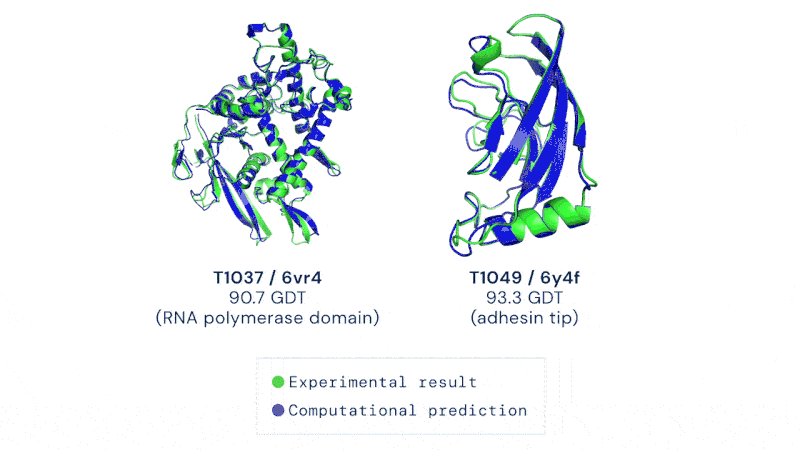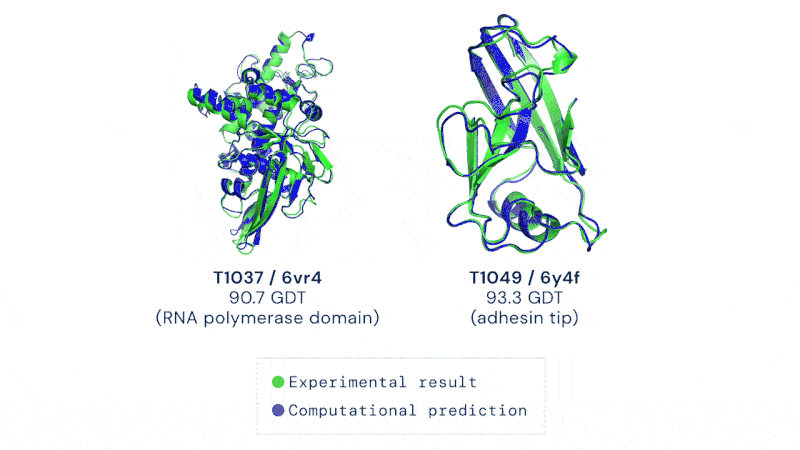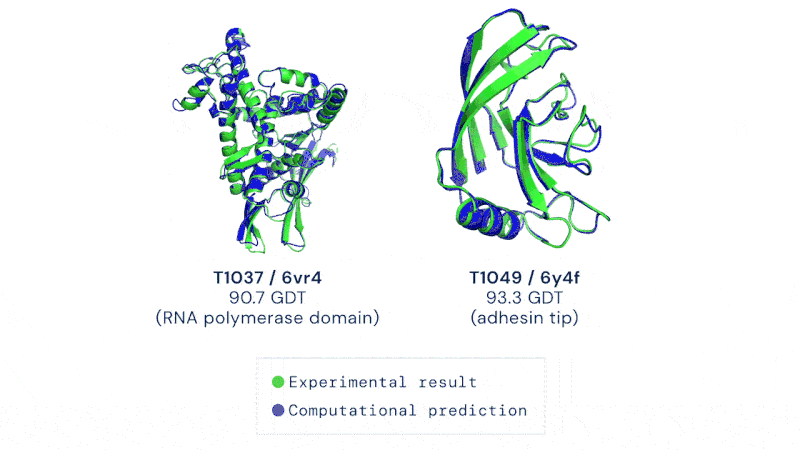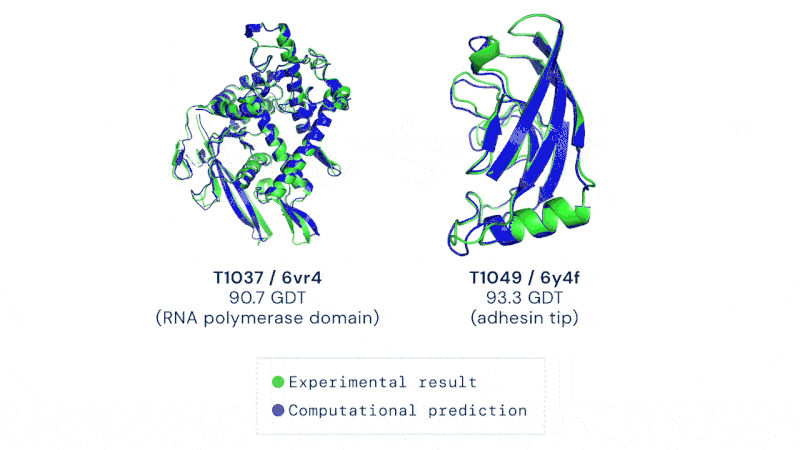
Having a method to predict this rapidly and without hard and extensive work could usher in a revolution in biology. It’s not just the development of new drugs and treatments, though that would be motivation enough. Development of enzymes that could break down plastic, biofuel production, even vaccine development could all be dramatically sped up by protein folding prediction algorithms.
Essentially, protein folding has become a bottleneck for biological research, and it’s exactly the kind of field where AI could make a big difference, unlocking new possibilities that seemed impossible even a few years ago.
At a more foundational level, mastering protein folding can even get us closer to understanding the biological building blocks that make up the world. Professor Andrei Lupas, Director of the Max Planck Institute for Developmental Biology and a CASP assessor, commented that:
“AlphaFold’s astonishingly accurate models have allowed us to solve a protein structure we were stuck on for close to a decade, relaunching our effort to understand how signals are transmitted across cell membranes.”
Why not everyone is convinced
The announcement of DeepMind’s achievements sent ripples through the science world, but not everyone was thrilled. A handful of researchers raised the point that just because it works in the CASP setting, doesn’t really mean it will work in real life, where the possibilities are far more varied.
Speaking to Business Insider, Max Little, an associate professor and senior lecturer in computer science at the University of Birmingham expressed skepticism about the real-world applications. Professor Michael Thompson, an expert in structural biology at the University of California, took to Twitter to express what he sees as unwarranted hype (see above), making the important point that the team at DeepMind hasn’t shared its code, and they haven’t even published a scientific paper with the results. Thompson did say “the advance in prediction is impressive.” He added: “However, making a big step forward is not the same as ‘solving’ a decades-old problem in biology and chemical physics.”
Lior Pachter, a professor of computational biology at the California Institute of Technology, echoed these feelings. It’s an important step, he argued, but protein folding is not solved by any means.
Just how big this achievement is remains to be seen, but it’s an important one no matter how you look at it. Whether it’s a stepping stone or a true breakthrough is not entirely clear at this moment, but researchers will surely help clear this out as quickly as possible.
In the meantime, if you want to have a deeper look at how AlphaFold was born and developed, here’s a video that’s bound to make you feel good:


