Science, mostly, progresses iteratively. But every now and then, a discovery will be made – often incidental to the main aim of the research – that is entirely unexpected. Such serendipitous findings enable us to leapfrog our usual incremental advances. They can even disrupt a whole field of research.
In 2003, the Human Genome Project, the 13-year-long international effort to fully sequence human DNA and identify all our genes, was completed. The Wellcome Sanger Institute, near Cambridge, England, was the only British organisation involved, completing the sequence of one-third of the genome.
While many of the great proclamations made at the launch of the project have yet to be realised, there is no doubt that sequencing the human genome was a technological game-changer for science in the way that, say, the invention of the printing press and microscopes were in previous centuries. And, just as for other disrupting technologies, genome sequencing has led to some wholly unexpected findings.
§
1. Chromosome shattering can cause cancer
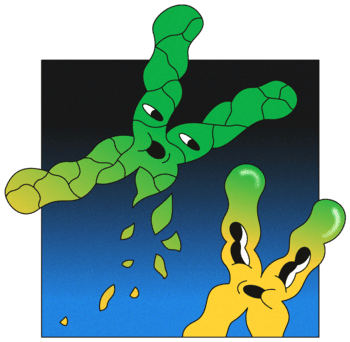
Cancer is a genetic disease. Through replication errors and mutations in DNA, healthy cells form tumours that can kill us. Today, researchers are sequencing the genomes of many tumours and through this transforming our understanding of what exactly cancer is.
Peter Campbell, who heads Sanger’s cancer programme, has made a few unexpected discoveries in his work with kidney tumours, but the most remarkable was an entirely new cancer-triggering mechanism.
He and his colleagues found that a chromosome can explode for unknown reasons, shattering into hundreds of pieces.
It was such a surprising finding that Campbell assumed it was because of a problem with the data. “Almost all of these things turn out to be rubbish in the data – someone’s mucked something up somewhere along the line. So usually it starts with us saying: we need to figure out what’s gone wrong,” he says. However, no obvious error could be found.
“The advantage of working with genetic data is it’s black and white, it behaves in a digital way – unlike, say, cells, which can look different on different days.”
Once he’d confirmed the discovery, Campbell allowed himself to get excited. Science, he notes, is usually a very long-term project: “You can be banging your head against problems that you can’t solve for weeks on end… then unexpected findings drop out all of a sudden.”
Campbell prepared himself for a backlash from the field. In the end, though, his research paper was received very positively, and others have since confirmed his findings, capturing the changes occurring in cells grown in the laboratory. The mechanism is likely to trigger many different cancers, researchers believe.
So has the finding shifted people’s thinking? “Yeah, I think so,” says Peter. “The best papers do that, but they come along not so often in one’s own career. We were lucky that we had very early access to the modern genetic technologies that allowed us to spot these patterns before others did.”
§
2. The Y chromosome is not useless after all
Haematologist George Vassiliou and his team were looking for new targets for drugs to treat leukaemia. One such target was a cancer-suppressing gene called UTX, which sits on the X chromosome.
As part of their investigations, they were using mice with faulty versions of the UTX gene to see when they got cancer. During these experiments, Vassiliou also tweaked a similar non-functional gene on the Y chromosome called UTY.
Aside from the gene that determines sex, Y chromosome genes were largely thought to be non-functional leftovers. In fact, the mole vole has evolved to lose its Y chromosome altogether.
Researchers have suggested that the same could happen in humans eventually. That was until Vassiliou’s team discovered that the UTY gene is functional in humans. What’s more, they found that it plays a significant role in suppressing cancer.
“It was very exciting,” he says. “At first, we were so happy to find another potential target for leukaemia treatment.”
But then there was a moment of realisation: “The Y chromosome actually does something! It is not useless after all.”
§
3. Bacterial genes we thought might not do anything actually affect how well vaccines work
Jukka Corander, a biostatistician at the Sanger Institute and University of Oslo, and Nicholas Croucher, a bacterial geneticist at Imperial College London, were exploring the genomics of bacterial infections. They used computer simulations of multiple strains to figure out what causes bacterial populations to change in unexpected ways after vaccination against them.
They were comparing the genomes of Streptococcus pneumoniae, a bacterium that causes severe illnesses such as pneumonia, sepsis and meningitis. This involved collections of bacteria from four different human populations around the world, three of which had been vaccinated against the bacterium.
All Streptococcus pneumoniae strains have around 2,000 genes. Three-quarters of these genes are very similar across strains. The remaining ‘accessory genes’ vary considerably between them. Bacterial strains can swap any gene through a process called horizontal gene transfer.
The modelling showed that the levels of accessory genes were very similar in the bacterial population before and after vaccination, even though the types of strain present had changed dramatically.
In other words, the strains that emerged after vaccination had similar sets of accessory genes to the strains eliminated by vaccination. So – far from being useless – accessory genes appear to play a role in how the bacterial population responds to a vaccine.
Importantly, some of the accessory genes that returned to their previous levels are involved in resistance to antibiotics.
“We had just published a paper that agreed with the ‘neutral model’ of the accessory genes,” Corander says, “so I was utterly astonished.”
This work provides the foundation for further research that will help predict which strains will spread most rapidly after a change to how we treat bacterial diseases.
“Without having access to high-definition genomes, we would never have seen this,” Corander says. “We wouldn’t even have known that so much variation exists in the genomes, let alone this important role of these rare accessory genes.”
Wellcome, the publisher of Mosaic, founded the Wellcome Sanger Institute in 1993 and has funded it ever since. The Sanger Institute celebrates its 25th anniversary in October 2018.
Nicholas Croucher holds a Sir Henry Dale Fellowship, which is funded by Wellcome and the Royal Society. Peter Campbell receives funding from Wellcome through a Senior Research Fellowship in Clinical Science.
This article was written by Gaia Vince and first appeared on Mosaic and is republished here under a Creative Commons licence.