An international team of researchers has imaged the surface of a bacterium in unprecedented detail, revealing in the process flaws in the outer membrane that acts as a barrier against antibiotics. These weak spots are like chinks in the bacteria’s armor that could one day be exploited by novel drugs to neutralize pathogens that are becoming increasingly resistant to conventional antibiotics.
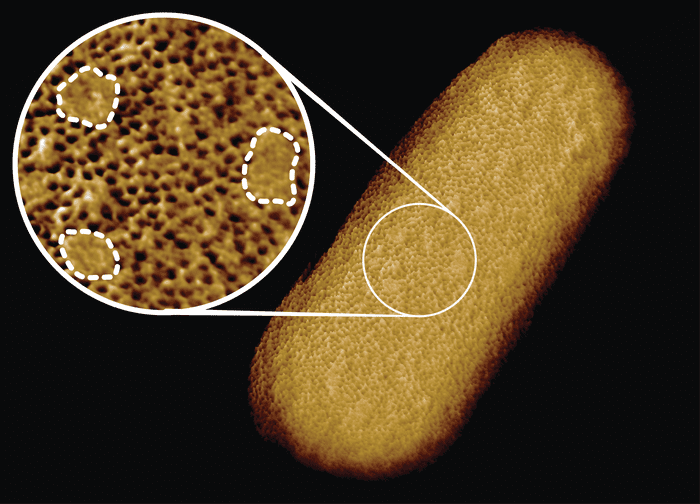
Patchy bacterium
Previously, scientists at the lab of Professor Bart Hoogenboom from University College London had used atomic force microscopy (AFM) to identify and pinpoint proteins on a bacterium’s surface. The method is a type of “feeling microscopy” by which a tiny sharp needle follows the contour of a sample similar to how a blind person may read Braille.
In their new research, first author and Ph.D. student Georgina Benn refined this process in order to better understand how some proteins in our blood manage to kill bacteria by punching holes into them. In doing so, she managed to image the entire surface of an Escherichia coli (E. coli) bacterium, at a staggeringly high resolution of less than five nanometers, or 1/10,000 of the thickness of the human hair.
This was a lot more challenging than it may sound. “You can’t just take your smartphone and take a few more pictures!” Hoogenboom said, adding that procedures had to be kept at a high standard every step of the way, from sticking bateria down to a glass substrate to various techniques related to the operation of the microscope so as not to destroy the sample.
“We needed to overcome many smaller obstacles to address a larger challenge, in our case that of reliably imaging bacteria with the sharpness as shown in this study and identifying what we were seeing,” Hoogenboom said.
“There is not one key breakthrough that allowed us to take this image; instead it resulted from the optimisation of a whole range of things that went into this. I like to compare this with cooking: An exquisite dish depends on a judicious choice and appropriate balance of several high-quality ingredients, rather than on just having added extra salt or pepper. And while I am of course biased, I think these images definitely deserve a Michelin star!”
E. coli belongs to a category of bacteria known as Gram-negative, characterized by a protective outer layer. Much to everyone’s surprise, this complete image showed that this protective shell wasn’t uniform but rather consists of a dense network of proteins interspersed with patches where there are no proteins. Instead, these patches are enriched with sugary molecules called glycolipids that help keep the outer membrane tight, but may also compromise the bacteria under certain conditions.
“We did very high-resolution microscopy on bacteria for a rather different purpose. We next, surprisingly, found that the architecture of the bacterial surface was much more patchy than previously thought, and not the homogeneous molecular brick wall that one might have expected. We also found that some types of patchiness correlated with enhanced vulnerability of the bacteria to some antibiotics. So we cautiously speculate that by understanding the patchy nature of the bacterial surface, we may learn how to kill a large class of a bacteria (so called Gram-negative bacteria) more effectively and thereby address health challenges related to antimicrobial resistance,” Hoogenboom told ZME Science.
These textbook-altering revelations may turn out to be formidable if they apply to other Gram-negative bacteria. Each year, just in the U.S., at least 2.8 million people contract an antibiotic-resistant infection, resulting in tens of thousands of deaths. Some bacteria can naturally resist certain kinds of antibiotics. Others can become resistant if their genes change or they get drug-resistant genes from other bacteria. The more drugs that we use and the more often, the less effective they are becoming against bacterial infections, which is why scientists across the world are looking for new tools to replace their dulling ones.
Besides the alternating patches on the surface of the bacteria, the researchers also observed a different type of pimple-like patch formed when parts of the membrane were flipped inside out due to mutations. These defects turned out to be sensitive to bacitracin, an antibiotic usually only effective against Gram-positive bacteria.
“We do not yet know if this can lead to new treatments. Quite generally, however, our study helps to better understand the architecture of the outer membrane, which is a main barrier that protects Gram-negative bacteria against antibiotics. So in the same way by which we can identify vulnerabilities in the structure of building when we know its detailed architecture, we anticipate that our findings may help us to identify vulnerabilities of bacteria that could be exploited by new antibiotics. Some of the patches we observe indeed appear to relate to enhanced vulnerability to some antibiotics, but this is definitely something that needs further exploration and study,” Hoogenboom said.
The patches aren’t a fluke of nature, though. In their study published in the Proceedings of the National Academy of Sciences, the researchers believe the alternating patches on the bacterial surface may explain how it can maintain its thick protective barrier while still allowing for furiously rapid growth. For instance, E. coli can double in size and then divide every twenty minutes, if given enough food. If the bacteria didn’t have the glycolipid patches, the membrane wouldn’t be able to stretch that easily.
However, before anyone gets too excited, these findings need to be replicated in other Gram-negative bacteria. The researchers hope to soon image Acinetobacter baumannii, a very common source of hospital infections that is becoming increasingly resistant to antibiotics.
“In spite of the high resolution we have got, there are many hidden aspects in our images, many molecules that we cannot directly identify. We would really hope to discover the full structure of the bacterial surface, a bit like the building guide for a Lego set, and then see how the effectiveness of various antibiotics depends on it,” Hoogenboom wrote in an email.
“As a scientist, I think the world is incredibly rich and even more so when we can look at it across many length scales. This can be by considering the very big (e.g., cosmology) or the very small (as in our study). It is just wonderful that we can “see” microbes that are undetectable by the human eye and even see the molecular-scale building blocks that they are made of.”
The research team also included scientists at National Physical Laboratory, King’s College London, University of Oxford and Princeton University.


