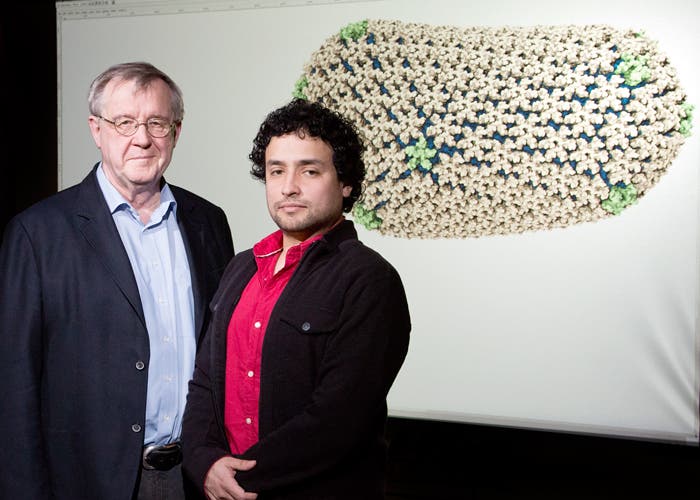
One big obstacle scientists face in their efforts to develop effective drugs against HIV is the virus’ capsid – an outer cell membrane-derived envelope and an inner viral protein shell that protects HIV essential proteins and genetic information. Current drugs have a hard time breaching this structure, however this might change. Using a supercomputer that crunched immense amounts of data, scientists have recently reported they have decoded the structure that contains and protects HIV’s genetic material.
“The capsid is critically important for HIV replication, so knowing its structure in detail could lead us to new drugs that can treat or prevent the infection,” said senior author Peijun Zhang, associate professor at the University of Pittsburgh School of Medicine.“This approach has the potential to be a powerful alternative to our current HIV therapies, which work by targeting certain enzymes, but drug resistance is an enormous challenge due to the virus’ high mutation rate.”
The capsid is one tricky fellow though, and accurately describing its structure was no easy task by any means. For one, the shell is comprised of nonuniform combinations of five- and six-subunit protein structures that link together to form an asymmetric shape. To model it, scientists had to piece together each of the 3 to 4 million atoms comprising it, while also accounting for all of the water molecules and salt ions also present, creating an output of some 64 million atoms.
The University of Pittsburgh researchers first used electron-scan microscope to see in incredible detail the protein molecules that comprise the capsid, then used an imaging technique to visualize how these molecules connect to each other to form the general shape of the shell. The data was then sent to University of Illinois physicists, who fed the data into computer models they ran on Blue Waters, their new supercomputer at the National Center for Supercomputing Applications capable processing 1 quadrillion operations per second. The data was run through the computer using a process called molecular dynamic flexible fitting, which outputted the minute details of the capsid’s structure.
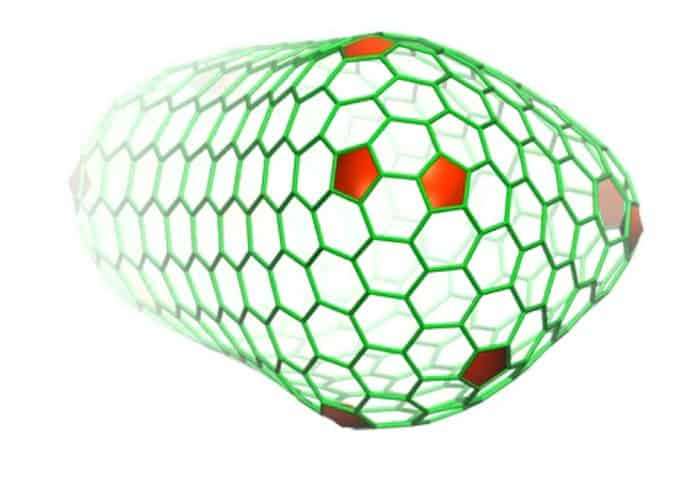
The process revealed a three-helix bundle with critical molecular interactions at the seams of the capsid, areas that are necessary for the shell’s assembly and stability, which represent vulnerabilities in the protective coat of the viral genome.
“This is a big structure, one of the biggest structures ever solved,” said University of Illinois physics professor Klaus Schulten. “It was very clear that it would require a huge amount of simulation – the largest simulation ever published. You basically simulate the physical characteristics and behavior of large biological molecules but you also incorporate the data into the simulation so that the model actually drives itself toward agreement with the data.”
If capsid assembly or disassembly is disrupted, viral replication, and consequently transmission, can be stopped. Now armed with this new found information, researchers have opened up a new front in their ongoing war with HIV.
“The capsid is very sensitive to mutation, so if we can disrupt those interfaces, we could interfere with capsid function,” Zhang said. “The capsid has to remain intact to protect the HIV genome and get it into the human cell, but once inside it has to come apart to release its content so that the virus can replicate. Developing drugs that cause capsid dysfunction by preventing its assembly or disassembly might stop the virus from reproducing.”
The findings appeared in Nature.






