Seahorses have a highly specialized morphology, with several characteristics unique in the animal kingdom. Now, researchers have found another remarkable trait of seahorses – their genome evolves extremely fast.

The genome project, comprises six evolutionary biologists from Professor Axel Meyer’s research team from Konstanz and researchers from China and Singapore. They chose the tiger tail seahorse Hippocampus comes. The assembled genome is 502Mb or about 1/6th the size of the human genome. The reason why they chose this species is pretty straightforward – just look at it.
It doesn’t look like any other fish – or any other creature for that matter – nor does it behave like any other creature. We don’t know if its features are adaptive or not, but we do know its evolutionary history to some extent. So scientists looked at 4,122 orthologous genes (segments of DNA with shared ancestry because of a speciation event) and calculated mutation rates, finding that the rate of change is extremely high.
Researchers believe that their study may partly explain why seahorses evolved the way they did.
“Our genome-wide analysis highlights several aspects that may have contributed to the highly specialized body plan and male pregnancy of seahorses,” they write in the study. “These include a higher protein and nucleotide evolutionary rate, loss of genes and expansion of gene families, with duplicated genes exhibiting new expression patterns, and loss of a selection of potential cis-regulatory elements.”
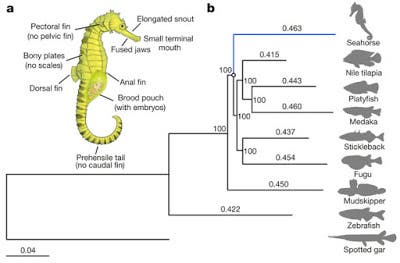
Seahorses possess one of the most highly specialized morphologies and reproductive behaviours. Their bodies include a toothless tubular mouth, a body covered with bony plates, a male brood pouch, and the absence of caudal and pelvic fins. Another surprising evolutive feature is male pregnancy. When mating, the female seahorse deposits up to 1,500 eggs in the male’s pouch. The male carries the eggs for 9 to 45 days until the seahorses emerge fully developed, but very small. After that, the male’s role is done but even so, it is a truly spectacular and almost unique trait (another type of fish does it – pipefishes).
Particularly noteworthy is the loss of the pelvic fins. Evolutionarily, you can consider them as analogous to human legs. The gene responsible for this feature (tbx4) is present in nearly all vertebrates, but is missing from the seahorse’s genome. Also, while some genes were absent, some genes were duplicated. When a gene is duplicated, it can fulfill a completely different role. In the seahorse, this is probably how a part of the newly created gene makes male pregnancy possible.
But not everyone is convinced. Writing on his website, Larry Moran, a Professor in the Department of Biochemistry at the University of Toronto, says that the accuracy of the study can’t be assessed without going deep into the big data algorithm they used.
“They looked at a set of 4,122 orthologous genes and calculated mutation rates. The results are shown in Figure 1 in the paper. The differences in distance are quite small, ranging from 94% tp 99% of the seahorse value (0.463) in the major clade. Nevertheless, the authors claim the difference is statistically significant. They also looked specifically at neutral changes and found the same thing—faster in seahorses. The implication is that the strange morphological differences between seahorses and other species of fish can be explained by a faster mutation rate.”
“Here’s the problem. I have no idea how they came up with these numbers. I can’t possibly evaluate the quality of their data to know whether it’s believable or not. Clearly the Nature referees thought it was good enough to publish. Those referees must be experts in this kind of analysis. Can someone out there help me understand the quality of this analysis?”
This indicates an issue with many modern studies. Even experts in respective fields can’t assess the validity of certain papers, because of the reliance on complex algorithms and big data. In other words, if you want to really understand the study, you often have to go through hundreds of lines of code (which are not readily accessible), in a language you are perhaps not familiar with – that’s simply not doable. Hopefully, reviewers are doing their job.
Journal Reference: Qiang Lin et al. The seahorse genome and the evolution of its specialized morphology. Nature, 2016; 540 (7633): 395 DOI: 10.1038/nature20595


