Scientists are pushing the frontiers of gene editing. A new study has just unveiled the first programmable RNA-guided system in eukaryotes. This system, based on a protein called Fanzor, uses RNA as a guide to precisely target DNA. It’s a lot like the now-famous CRISPR system, which is mainly found in bacteria.
The implications of this finding are far-reaching, as it opens up new possibilities for more effective and efficient gene editing techniques.
A New Frontier in Gene Editing
The CRISPR-Cas system, which has garnered significant attention in recent years, was first discovered in prokaryotes, which include bacteria and other single-cell organisms lacking nuclei.
CRISPR, which stands for Clustered Regularly Interspaced Short Palindromic Repeats, is a natural defense mechanism found in bacteria and archaea. It acts as a molecular “scissors” that can precisely edit DNA. It consists of two main components: a guide RNA molecule and a protein called Cas9.
The guide RNA acts as a homing device, guiding the Cas9 protein to a specific location in the genome that matches the RNA sequence. Once the Cas9 protein reaches its target, it cuts the DNA strand, allowing researchers to modify or replace the genetic material extremely precisely. It’s like cutting and pasting genes, so one can easily imagine how impactful and powerful this method can be.
CRISPR has the potential to correct genetic mutations responsible for inherited diseases, such as cystic fibrosis and sickle cell anemia. It can enhance crop yields, improve nutritional content, and make plants more resistant to pests, diseases, and adverse environmental conditions. Some scientists are even contemplating using CRISPR to resurrect long-extinct species. Indeed, some are already actively working to resurrect the wooly mammoth, for instance.
But despite all its bells and whistles, CRISPR isn’t perfect.
Scientists have long wondered whether similar systems exist in eukaryotes, which comprise fungi, plants, and animals — including us humans. This latest study led by Feng Zhang and his team at the prestigious McGovern Institute for Brain Research at MIT provides compelling evidence that RNA-guided DNA-cutting mechanisms are indeed present across all kingdoms of life.
The Fanzor system
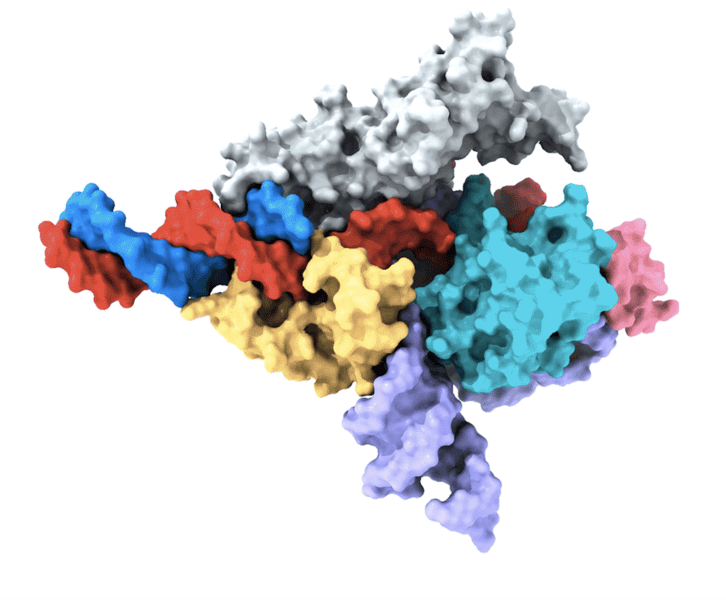
Several years ago, the team discovered a class of RNA-programmable systems in prokaryotes known as OMEGAs, which share similarities with the Fanzor proteins found in eukaryotes. This observation hinted at the possibility that Fanzor enzymes might also utilize an RNA-guided mechanism to target and cut DNA.
In their latest study, the researchers isolated Fanzor proteins from various organisms, including fungi, algae, and amoebae, as well as a clam species called the northern quahog. Co-first author Makoto Saito led the biochemical characterization of the Fanzor proteins, demonstrating that they are endonuclease enzymes capable of cutting DNA.
These proteins utilize non-coding RNAs called ωRNAs (OMEGA RNAs) to target specific sites in the genome. This marks the first time such a mechanism has been identified in eukaryotes, particularly animals.
Unlike CRISPR proteins, Fanzor enzymes are encoded within the eukaryotic genome, residing in transposable elements.
Phylogenetic analysis conducted by the team suggests that Fanzor genes have been transferred from bacteria to eukaryotes through a phenomenon known as horizontal gene transfer. In contrast to vertical gene transfer, which occurs during reproduction from parent to offspring, horizontal gene transfer involves the transfer of genetic material between organisms of the same or different species.
These ancient OMEGA systems, which actually predate CRISPR, are abundant in nature and have the ability to migrate between prokaryotes and eukaryotes. The implication is that Fanzor proteins can be used in the same role as CRISPR proteins to guide genetic changes with surgical accuracy.
“CRISPR-based systems are widely used and powerful because they can be easily reprogrammed to target different sites in the genome,” said Zhang.
“This new system is another way to make precise changes in human cells, complementing the genome editing tools we already have.”
Unleashing the Editing Potential
To explore the genome editing capabilities of the Fanzor system, the researchers conducted experiments on human cells. The results demonstrated that Fanzor can introduce insertions and deletions at targeted sites within the genome.
Although initially less efficient than CRISPR-Cas systems in cutting DNA, the Fanzor system’s effectiveness was enhanced through systematic engineering. The team introduced specific mutations into the protein, resulting in a tenfold increase in its activity.
Additionally, unlike some CRISPR systems and the OMEGA protein TnpB, the Fanzor protein derived from fungi did not exhibit collateral activity, where the enzyme cleaves its DNA target along with degrading nearby DNA or RNA. This finding suggests that Fanzors could potentially be developed into efficient, safer genome editors.
Another co-first author of the study, Peiyu Xu, focused on analyzing the molecular structure of the Fanzor/ωRNA complex. Through this analysis, Xu discovered that Fanzor shares structural similarities with its prokaryotic counterpart, the CRISPR-Cas12 protein. However, the interaction between the ωRNA and the catalytic domains of Fanzor is more extensive, indicating that the ωRNA may play a role in the catalytic reactions.
Finally, Fanzor systems are more compact than CRISPR proteins, which means they can be more easily delivered to cells and tissues.
| CRISPR | Fanzor |
|---|---|
| Discovered in prokaryotes (bacteria and single-cell organisms) | Discovered in eukaryotes (organisms including fungi, plants, and animals) |
| Relies on the Cas proteins for targeting and cutting DNA | Relies on the Fanzor proteins for targeting and cutting DNA |
| Uses CRISPR RNA (crRNA) as a guide to identify target sequences | Uses non-coding RNA called ωRNAs as a guide to target specific sites in the genome |
| Can be easily reprogrammed to target different sites in the genome | Can also be easily reprogrammed to target specific genome sites |
| Generally more efficient in cutting DNA compared to the initial efficiency of Fanzor | Through systematic engineering, the activity of Fanzor can be enhanced, making it more efficient in cutting DNA |
| Collateral activity may occur, where the enzyme cleaves its DNA target along with degrading nearby DNA or RNA | Fanzor derived from fungi does not exhibit collateral activity, making it more precise in its editing |
| Widely used and established as a genome editing tool | Holds potential as a powerful new genome editing technology, complementing existing tools |
A Promising Future
Similar to CRISPR-based systems, the Fanzor system can be reprogrammed to target specific sites within the genome. This flexibility positions it as a potentially powerful tool for genome editing in research and therapeutic applications. And since there is an abundance of RNA-guided endonucleases, such as Fanzors, throughout various kingdoms of life, it is highly likely that additional RNA-programmable systems remain to be discovered.
“Nature is amazing. There’s so much diversity,” says Zhang. “There are probably more RNA-programmable systems out there, and we’re continuing to explore and will hopefully discover more.”
The findings appeared in the journal Nature.






