Digital storage has gone a long day since the advent of digital computing. It used to take hard drives the size of a boat to store a fraction of the data you can now access with a smartphone. The bottleneck is closing in though and the progress in increasing storage density is incremental while demand is soaring. Silicon, a faithful ally, seems to be reaching its limits, so scientists are looking for alternatives. For archiving purposes, at least, DNA — the blueprint or genetic recipe that codes all life — might be worth considering. One team, for instance, coded digital images into synthetic DNA using a novel method, then decoded and read this data.

We’ve known DNA is an amazing storage medium ever since it was first discovered, after all only one cell contains all the information necessary to build an entire human being. It’s actually fantastically good at storing data since the information can be coded in a volume unlike a planar surface as is the case for man-made flash and hard drives.
In one single cubic millimeter, DNA can hold up to 1 exabyte. Only 4 grams of DNA can hold up to all the digital data we create annually. This is much denser than digital storage media such as flash drives, and more stable since the DNA sequences could be read thousands of years after they were encoded.
“A large part of building better computers is about finding better materials to build computers with,” says Luis Ceze, an associate professor in the Computer Science Department at the University of Washington. “So, silicon happens to be a fantastic material, but it’s reaching a point where it’s unclear that we can continue pushing forward with silicon. So I find it fascinating that biology has evolved many molecules that are useful for building better computers in the future.”
Ceze and colleagues first start by ‘translating’ data from digital 1s and 0s into the four letters of DNA: A, G, C and T. A DNA synthesizer creates short strands of DNA that each holds a part of a file’s code. A DNA sequencer can then read and retrieve the information encoded, akin to the way scientists use sequencing today to read genetic information about our ancestors. To demonstrate, they encoded a couple of images into DNA, then retrieved and displayed the data.
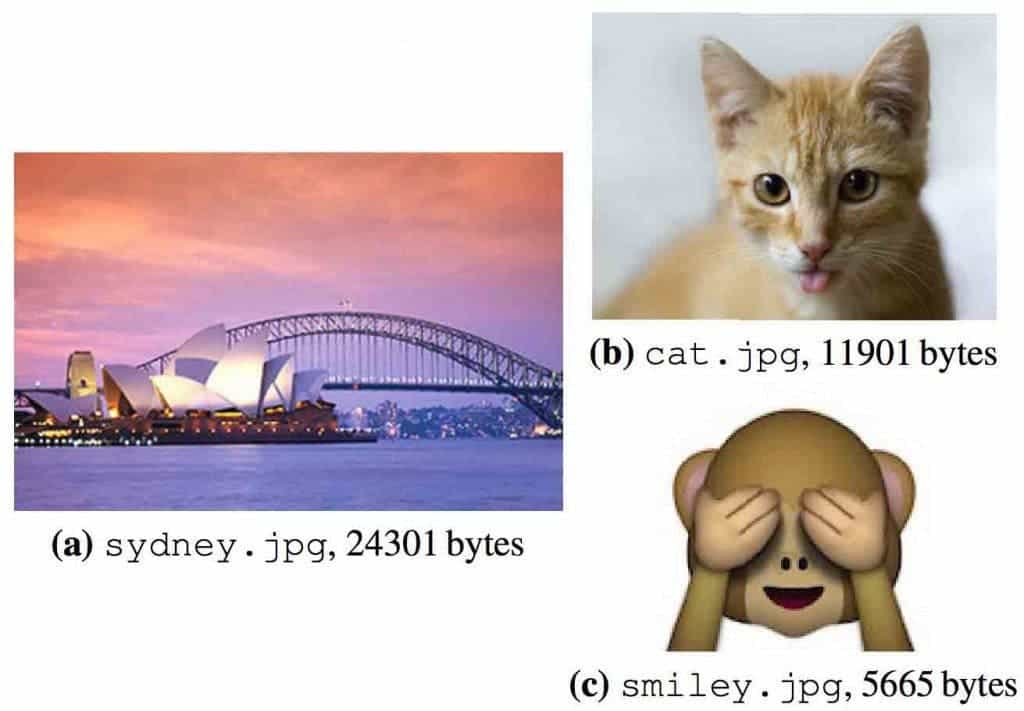
Ceze and team aren’t the first to encode data into DNA. One notable attempt from 2012 encoded a book which was 53,400 words long into DNA sequences, along with 11 images in JPG format and a JavaScript program. This most recent research is more efficient since it reduces redundancies. Fewer strands of DNA have to be synthesized to account for errors, significantly improving storage. Concerning retrieval, data can be read without actually sequencing all of the DNA strands.
We might be a long way from using DNA inside your computer, though. Unlike flash drives, DNA has to be read by shuttling molecules around. This process is at snail’s pace compared to the movements of electrons which move with the speed of light. Instead, someday DNA will be used to first store sensitive information for archival purposes. It won’t be alone either. Previously, researchers crammed 360TB worth of five dimensional (5D) digital data onto a small quartz disk, which should be stable for 13 billion years.






