The human genome was sequenced for the first time in 2003, tremendously influencing genetics and biological research ever since. But despite huge leaps in our understanding of the human blueprint, these early efforts were not complete. It wasn’t until 2022 that scientists filled the gaps and sequenced the complete human genome — and we’re still not done yet.
Now, a consortium of scientists has raised the bar once more by publishing the first human “pangenome”, which incorporates the genomes of 47 individuals from across the world.
By combining the genetic material of 47 individuals from diverse ancestral backgrounds, this revolutionary reference provides an unparalleled understanding of the intricacies of human genetic diversity.
In an extraordinary series of papers published today in prestigious scientific journals, this game-changing achievement promises to reshape the landscape of genomics, delivering profound insights into our shared human heritage.
Unlocking the Secrets of Genetic Diversity: The Human Pangenome Revolution
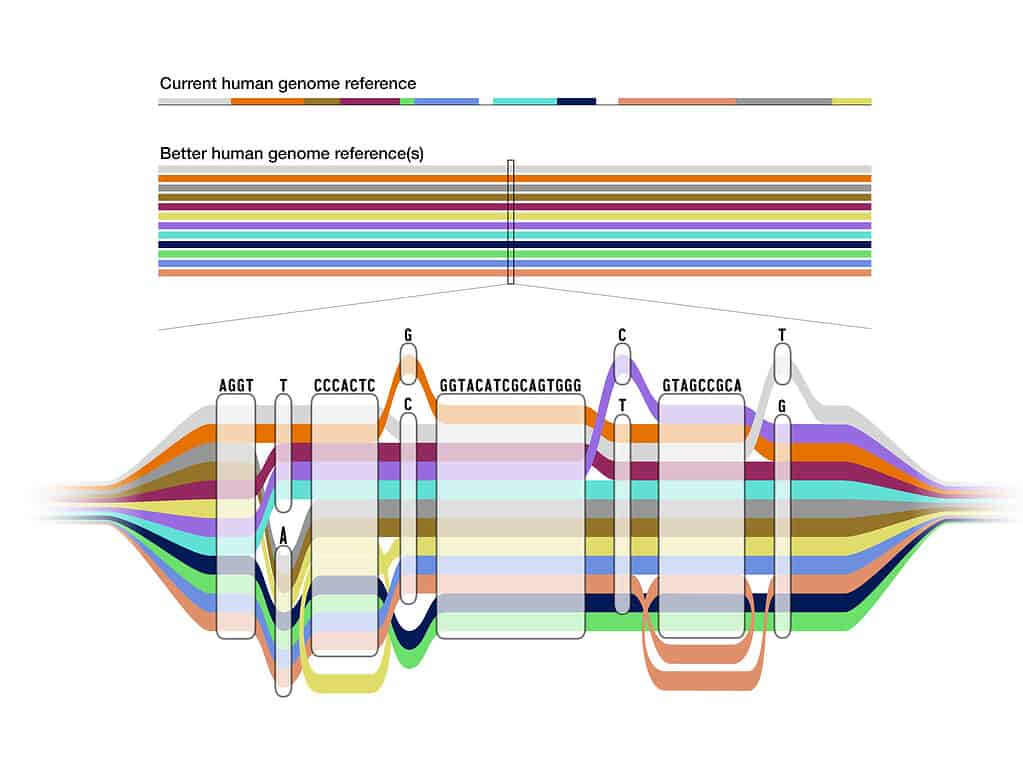
Although 99.9% of the genome is the same from person to person, there is a lot of diversity found in that final 0.1%. Even with the complete human genome that scientists published last year, 70% of the sequence scientists use to benchmark genetic variation still comes from a single person.
Needless to say, this is a problem for genetic research. This limitation introduces a phenomenon known as reference bias, hindering our ability to comprehensively analyze genomes.
Enter the pangenome. Unlike its predecessor, the pangenome embraces diversity and inclusivity, blending the genomes of 47 individuals from diverse ancestral backgrounds.
Visualize it as a tapestry of human variation, where common genetic sequences form a seamless path while diverging areas unveil the unique genetic footprints of different populations.
With the addition of 119 million DNA bases—those fundamental “letters” that form our genetic code—the pangenome transcends the limitations of a single reference genome.
In the process, the pangenome offers unparalleled accuracy, completeness, and, most importantly, an extraordinary ability to uncover genetic variants that have previously eluded our grasp.
A New Genomic Frontier Unveiled
Genomic variation comes in various forms—ranging from subtle differences in individual DNA bases to larger structural variants that span 50 base pairs or more. These structural variants can profoundly impact our health.
However, until now, our ability to identify them has been severely limited. A mere 30% of these structural variants have been detectable with existing technologies and the constraints of a single reference genome.
Within the 119 million new bases added to the pangenome, approximately 90 million derive from structural variations. These include inversions, insertions, deletions, and tandem repeats (segments of genetic material repeated multiple times).
These additional bases open doors to uncharted territories of the genome, shedding light on regions that previously lacked reference, and potentially unraveling associations between structural variants and diseases such as autism, schizophrenia, immune disorders, and coronary heart disease.
The implications of this breakthrough extend far beyond structural variants. When it comes to identifying smaller genetic variations, such as single-base changes, the pangenome outshines its predecessor with a 34% increase in accuracy.
By harnessing the vast amount of data present in the pangenome, scientists can now uncover these minute variations with unparalleled precision.
Mapping the Road to Inheritance
Within each of us lies a paired set of chromosomes—one inherited from our mother, the other from our father. The pangenome is now helping scientists unravel this complex web of inheritance.
With the power of haplotype resolution, the pangenome confidently distinguishes between the two sets of parental chromosomes, something that used to be extremely challenging before—a remarkable scientific achievement. This newfound knowledge empowers scientists to delve deeper into the mysteries of gene inheritance and the role it plays in various diseases.
This also means that the pangenome encompasses not just one but an astounding 94 distinct genome sequences. And the journey doesn’t end there. By 2024, the researchers plan to expand this collection to include 700 reference genomes, encompassing an even broader spectrum of human genetic diversity.
A Tapestry Woven with Precision
Behind the scenes, a symphony of computational techniques and cutting-edge algorithms has brought the pangenome to life through the Human Pangenome Reference Consortium (HPRC). Dozens of researchers from many institutions across the US, UK, and Germany participated in this landmark achievement.
For instance, the UC Santa Cruz Computational Genomics lab, led by Benedict Paten, has spearheaded the development of advanced methods to align multiple genome sequences into a unified structure—an intricate pangenome graph. Within this graph, shared paths represent regions of similarity, while diverging paths highlight areas of genetic variation.
These paths are meticulously crafted, ensuring that each genome within the pangenome reference attains exceptional quality and accuracy.
“The draft pangenome is an important proof of principle that we hope is going to influence a lot of people and get them thinking about the pangenome and how it might affect their work,” Paten said.
“Looking ahead, we see a lot of engagement with other groups—it takes a lot of different people to build something that is going to become a big community resource.”
All of the 47 diploid genomes were sourced from volunteers who participated in the 1000 Genomes Project (1000G) and agreed to share their anonymized genetic sequences in publically available databases. These openly consented samples—sourced from diverse backgrounds—pave the way for unrestricted access to this invaluable resource without the privacy barriers that typically accompany genome research.
Surely, this is just the beginning.
Overall, by incorporating the genomes of dozens of individuals from around the world, the pangenome provides a far richer source of data than the original reference genome.
“We are introducing more diversity and equity into the reference by sampling diverse human beings and including them in this structure that everyone can use,” said Paten, who is the senior author on the main marker paper. “One genome isn’t enough to represent everybody—the pangenome will ultimately be something that is inclusive and representative.”
The new findings were described in four separate papers published today in the journal Genome Research, Nature, Nature Biotechnology, and Nature Methods.
- Benedict Paten et al, A draft human pangenome reference, Nature Biotechnology (2023). DOI: 10.1038/s41586-023-05896-x. www.nature.com/articles/s41586-023-05896-x
- Vollger et al, Increased mutation rate and gene conversion within human segmental duplications, Nature (2023). DOI: 10.1038/s41586-023-05895-y
- Guarracino et al, Recombination between heterologous human acrocentric chromosomes, Nature (2023). DOI: 10.1038/s41586-023-05976-y
- Hickey et al, Pangenome graph construction from genome alignment with minigraph-cactus, Nature Biotechnology (2023). DOI: 10.1038/s41587-023-01793-w






