Using an algorithm designed to stream videos on mobile phones, researchers showed how to maximize the data storage potential of DNA. Their method can encode 215 petabytes of data — twice as much as Google and Facebook combined hold in their servers– on a single gram of DNA. This is 100 times more than previously demonstrated. Moreover, the researchers encoded an operating system and movie onto the DNA were able to successfully retrieve the data from sequenced DNA without any errors.

Every day, we create 2.5 quintillion bytes of data, and at an ever increasing pace. IBM estimates 90% of the data in the world today has been created in the last two years alone. As more and more of our lives gets transcribed in digital form, this trend will only get amplified. One problem is that hard drives and magnetic tapes will soon become inadequate for storing such vasts amount of ‘big data’ — which is where DNA comes in.
It’s often called the ‘blueprint of life’, for obvious reasons. Every cell in our bodies, every instinct, is coded in base sequences of A, G, C and T, DNA’s four nucleotide bases. Ever since DNA was first discovered in the 1950s by James Watson and Francis Crick, scientists quickly realized huge quantities of data could be stored at high density in only a few molecules. Additionally, DNA can be stable for a long time as a recent study showed which recovered DNA from 430,000-year-old human ancestor found in a cave in Spain.
“DNA won’t degrade over time like cassette tapes and CDs, and it won’t become obsolete — if it does, we have bigger problems,” said study coauthor Yaniv Erlich, a computer science professor at Columbia Engineering, a member of Columbia’s Data Science Institute
Erlich and colleagues at the New York Genome Center (NYGC) chose six files to write into DNA: a full computer operating system, an 1895 French film, “Arrival of a train at La Ciotat,” a $50 Amazon gift card, a computer virus, a Pioneer plaque and a 1948 study by information theorist Claude Shannon.
All the files were compressed into a single master file then split into short strings of binary code, all 1s and 0s. The researchers used a technique called fountain codes which Erlich remembered from graduate school, to make the reading and writing process more efficient. Using the algorithm, they packaged the strings randomly into ‘droplets’, where each 1 and 0 was mapped to one of the DNA’s four nucleotide bases (A,G,C,T). The algorithm proved essential to store and retrieve the encoded data since it corrects and deletes letter combinations known to provoke errors.
Once they finished, they ended up with a huge text file made of 72,000 DNA strands each 200 bases long. The text file was sent to a startup from San Francisco called Twist Bioscience which turned all that digital data into biological data by synthesizing DNA. Two weeks later, Erlich received a vial containing DNA which encoded all of his previous work.
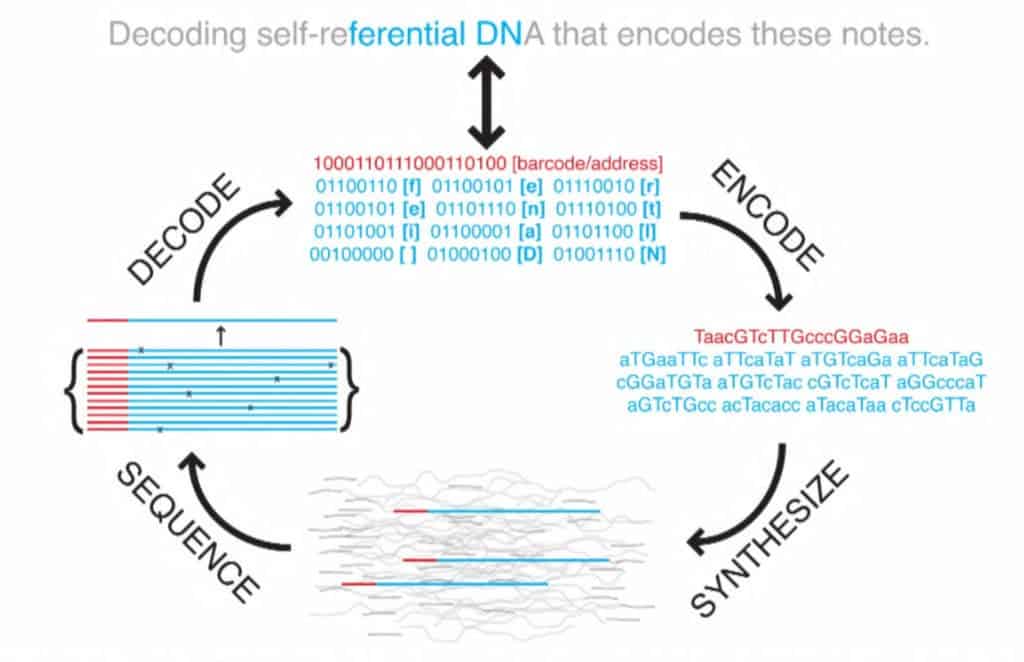
To retrieve the files, the researchers used common DNA sequencing tools as well as a special software which translates all the As, Gs, Cs, and Ts back into binary. The whole processes worked flawlessly seeing how the data was retrieved with no errors.
To demonstrate, Erlich installed on a virtual machine the operating system he had encoded in the DNA and played a game of Minesweeper to celebrate. Chapeau!
“We believe this is the highest-density data-storage device ever created,” said Erlich.
Erlich didn’t stop there. He and colleagues showed that you could copy the encoded data as many times as you wish. To copy the data, it’s just a matter of multiplying the DNA sample through polymerase chain reaction (PCR). The team showed that copies of copies of copies could have their data retrieved as error-free as the original sample, as reported in the journal Science.

There are some caveats, however, which I should mention. It cost $7,000 to synthesize the DNA and another $2,000 to read it. But it’s also worth keeping in mind that sequencing DNA is getting exponentially cheaper. It cost us $2.7 billion and 15 years of work to sequence the first human genome, then starting from 2008 the cost came down from $10 million to the couple-thousand-dollar mark. Sequencing DNA might become as cheap as running electricity through transistors at some point in the not very distant future.
Another thing we should mention is that DNA storage isn’t meant for mundane use. As it stands today, you can’t replace your HDD with DNA in a home computer for instance because the read-write time can take days. Instead, DNA might be our best solution for archiving the troves of data that are amounting to insane quantities with each passing day. And who knows, maybe someone can find a way to code and encode data in molecules as fast as electrons zip through a transistor — but that seems highly unlikely, if not impossible.


