CRISPR-Cas9 technology revolutionized gene editing, enabling scientists to edit parts of the genome by removing, adding or altering sections of the DNA sequence. Since the tool was first mentioned in 2002, over 4,800 articles employing CRISPR appeared PubMed, with exponential adoption rates over the last three years. But CRISPR isn’t perfect and due to so-called ‘off-target effects’, the approach can sometimes cause undesirable mutations. You never want to hear ‘Oops!’ when altering DNA. Now, two new studies have tuned gene editing up a notch by enabling scientists to target and change a single letter in a string of DNA bases. No molecular scissors required.

Instead of slicing through the two spiraled strands of bases — the famous DNA double-helix — American researchers at the Broad Institute and MIT, on one hand, and at Harvard University, on the other hand, targeted DNA’s close chemical cousin: RNA. The two are different both structurally and functionally. DNA, for instance, is comprised of deoxyribose sugars rather than ribose sugars, as is the case of RNA. DNA is a sort of genetic blueprint responsible for storing and transferring genetic information. RNA, on the other hand, acts like a secondary copy which directly codes for individual amino acids. If they could precisely change individual letters in a RNA sequence, the scientists reckoned, then it would be possible to reverse disease-causing mutations.
Led by David Liu, the Harvard team presented a new gene editing technique called ‘base editing’ in the journal Nature. If CRISPR is like using a pair of molecular scissors to splice DNA and alter a genome, then base editing is like using a pencil and eraser, Liu says. In other words, this is a far sharper way of altering genomes in some applications. CRISPR will still have an important role to play for years to come.
The typo that can kill
The rules of base pairing (or nucleotide pairing) dictate that A (the purine adenine) always pairs up with T (the pyrimidine thymine) and C (the pyrimidine cytosine) always pairs with G (the purine guanine). What Liu and colleagues managed to do is they engineered an enzyme that targets the “A” base and changes it into a base called inosine, which is read as as guanine, the “G.” Then, the cell’s DNA repair machinery tries to ‘fix’ the complementary strand of DNA across the gap where the T is by inserting a C. What you eventually get is an A-T to G-C conversion.
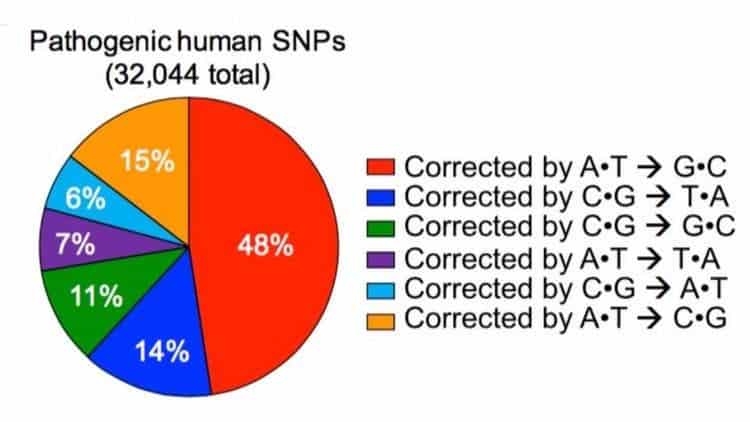
This sort of letter switching happens all the time in nature and regularly cause disease in humans. A mutation from G to A, which is very common, has been implicated in cases of focal epilepsy, Duchenne muscular dystrophy, and Parkinson’s disease. According to Liu, about half of the 32,000 known pathogenic point mutations in humans can be traced down to mutations that change G-C to an A-T.
The second paper published in Science and authored by scientists at the Broad Institute and MIT took a different route for A-G conversion. They inserted a different adenosine deaminase enzyme into Crispr-Cas13, which is a variant genome editor that works on RNA. The enzyme used in this case, called PspCas13b, comes from the Prevotella bacteria and proved to be the most effective at inactivating RNA. The new CRISPR-based system is called RNA Editing for Programmable A to I Replacement, or “REPAIR.”
Senior author Feng Zhang says REPAIR has the ability to reverse the impact of any pathogenic G-to-A mutation regardless of its surrounding nucleotide sequence, with the potential to operate in any cell type. What’s more, the changes are reversible unlike the permanent alterations implied by DNA editing.
“REPAIR can fix mutations without tampering with the genome, and because RNA naturally degrades, it’s a potentially reversible fix,” explained co-first author David Cox, a graduate student in Zhang’s lab.
Both approaches provide a sharper and, most importantly, now reversible approach to gene editing. The main problem with CRISPR-Cas9 doesn’t lie in cutting but in the repair — the addition and tossing of bases which can sometimes lead to unwanted mutations. Operating on RNA, however, circumnavigates this issue because RNA repair doesn’t involve indels (stochastic insertions or deletions). With the scariest outcomes of CRISPR now potentially gone, gene therapy could grow immensely. Genetic diseases due to base defects could be eradicated and new classes of drugs could surface. Before that can happen, researchers need to find the best way to deliver the base editor machinery to the right tissues in the body and into the right cells. Scientists have a tremendous amount of work ahead of them but the future already looks intense.
“There’s immense natural diversity in these enzymes,” said co-first author Jonathan Gootenberg, a graduate student in both Zhang’s lab and the lab of Broad core institute member Aviv Regev. “We’re always looking to harness the power of nature to carry out these changes.”






