The molecular mechanisms underpinning the spread of drug resistance in bacteria populations have been identified — and a new class of molecules has been designed to fight it.
The rise of multi-drug resistant bacteria is often — and to my mind, as well as the WHO’s — rightly held to be one of the biggest current threats to global health. A large part of what constitutes this threat is that bacteria can share resistance among themselves — like IT guys swapping USB sticks with new firewall software, bacteria can share genes encoding antibiotic resistance.
In a bid to nip this growing threat in the bud, researchers at the European Molecular Biology Laboratory (EMBL) have uncovered and then unraveled one of the major resistance-transfer mechanisms. They’ve also developed proof-of-concept molecules to carry out this bacterial sabotage.
Resisting the resistance
Over time, bacteria have developed a certain resistance level to most drugs we use today. The worst are arguably those that have developed resistance to multiple classes of antibiotics; examples range from MRSA (methicillin-resistant Staphylococcus aureus), VRE (vancomycin-resistant enterococcus), and ESBL (extended spectrum beta-lactamase) producing Enterobacteriaceae.
One of the major drivers of resistance spread throughout bacterial populations are transposons. Also called ‘jumping DNA’, they are bits of genetic code that can autonomously move throughout the genome. When this movement occurs between bacteria, it spreads antibiotic resistance genes among individuals. Very bad for us.
Under the leadership of Orsolya Barabas, one research team at the EMBL became the first to determine the structure of a crystal-like, protein-DNA structure which inserts these transposons in recipient bacteria. Dubbed the transposase protein, this molecule could hold the key to throwing the whole process into disarray.
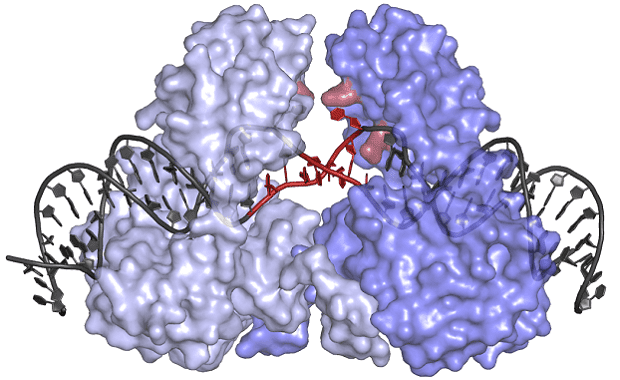
Image credits Cell.
The protein has an unusual shape, which allows it to bind to DNA in an inactive state, keeping the transposon safe from potential chemical or physical damage until it’s delivered to its new host. Its shape also forces the transposon DNA to unwind, the team notes, allowing the protein to insert these genes into a wide array of locations within the genomes of many different bacteria.
“If you think of ropes or wires, they are usually bundled and wound-up to make them stronger. If you want to tear or cut one, it’s much easier if you unwind and loosen it first,” says EMBL group leader Orsolya Barabas, who led the work.
“It’s the same for DNA, and the transposon transfer mechanism takes advantage of this.”
Because the transposase protein first unwinds and separates the transposon’s strands, they can more readily be cut and pasted to a new site in the recipient genome. Again, very bad for us.
Luckily, Barabas’ team used the protein’s crystal structure to develop molecules that should block the transposons’ movement through two mechanisms. The first prevents the transposase protein from activating by blocking its architecture with a newly designed peptide, a short chain of amino acids — in other words, it wedges itself in the transposase so that it can’t unfurl and deliver the DNA cargo.
The second method ‘corrupts’ the genetic data. This molecule, a DNA-mimic, binds to the transposon and blocks the DNA strand replacement in the host; no replacement, no resistance transfer.
“As we believe these features are broadly present in these jumping DNA elements, but not in related cellular systems, they may be quite specific to transposons. This way, we can target only the bacteria we want, and not the many good bacteria in our bodies and the environment,” Barabas explains.
The molecules are still far from trials with living hosts. For now, Barabas and her colleagues will focus on better understanding the transfer mechanisms, as well as on developing and testing new strategies to block it.
The paper “Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance” has been published in the journal Cell.


