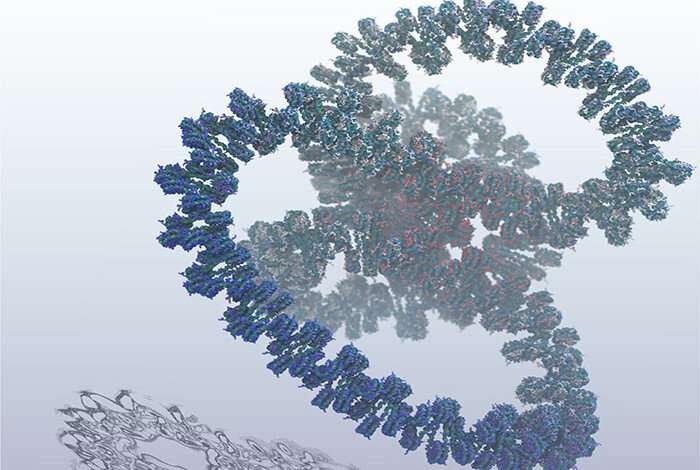
Researchers have simulated a billion atoms which make up an entire human gene for a split-second. This is the largest simulation of human DNA and an important milestone towards the ultimate goal of digitally reproducing the human genome.
“It is important to understand DNA at this level of detail because we want to understand precisely how genes turn on and off,” said Karissa Sanbonmatsu, a structural biologist at Los Alamos. “Knowing how this happens could unlock the secrets to how many diseases occur.”
Sanbonmatsu and colleagues performed their study on the Los Alamos’ Trinity supercomputer, the sixth fastest in the world. But even for this behemoth, simulating the intricate complexities of DNA was a huge challenge that required all of its computing resources. The model is quite slow too, simulating just one nanosecond of molecular activity per day.
DNA, or deoxyribonucleic acid, is the hereditary material in humans and almost all other organisms. This incredible molecule contains all the instructions an organism needs to develop, live, and reproduce. Its structure is so neatly compacted and precise that you could string together all the DNA in a human body to wrap around the earth 2.5 million times.
The reason why the blueprint for life is so compact has to do with the way the string-like molecule is wound up in a network of tiny spools. The various ways in which these spools wind and unwind turn genes on and off. In other words, when the DNA is more compacted, genes are turned off and when DNA expands, genes are turned on.
Researchers do not yet fully understand how all of this process pans out, which is why they’ve developed this atomistic model. Solving this mystery could one day lead to novel gene therapies and medical applications.
But before that happens, we need much faster computers. Modeling billions and billions of atoms all moving at the same time requires phenomenal resources. And, if we want to model an entire chromosome (or even the human genome), scientists will have to wait for the next generation of supercomputers, such as exascale computers, which will be many times faster than today’s machines.
Scientific reference: Jaewoon Jung et al. Scaling molecular dynamics beyond 100,000 processor cores for large‐scale biophysical simulations, Journal of Computational Chemistry (2019). DOI: 10.1002/jcc.25840.






