Tumor suppressor genes are normally busy keeping your cell’s growth cycle regulated, and when in good working order, prevent cells becoming malignant. A new study finds that many cancers are caused by mutations that block the tumor suppressor gene’s effect, through a process called Intron Retention. Introns are normally removed after a gene is transcribed into RNA, but in intron retention, one is accidentally left in place. The result can be disastrous, leading to cancer and possibly other disease.

Introns are found in complex cells, like those of animals and plants, but not in simple cells like bacteria. Introns are non-coding sequences, meaning that even though the intron is part of the gene, it’s DNA is not used in the gene’s instruction for making protein. Within the gene, the introns are placed between exons – the sequences of DNA that are the actual code for protein. A single gene may contain many introns and exons. When a gene becomes activated, the cell transcribes the DNA into messanger-RNA (mRNA), which then leaves the cell nucleus to be translated into a protein by ribosomes in the cytoplasm. Under normal conditions, both intron and exon DNA on the gene get transcribed into mRNA, but the intron is then cut out of the mRNA prior to leaving the cell nucleus, and so never goes on to the ribosome. Therefore, the mature mRNA comes only from the DNA of the gene’s exons – all unwanted introns having been removed. The removal of the introns from the mRNA is called splicing, and is carried out by complex cellular machinery, composed of both protein and RNA, called the Splicosome.
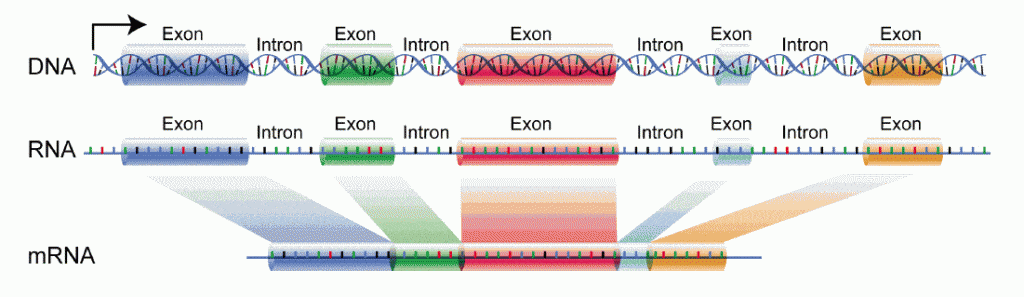
Splicing is an important invention of complex cells, leading to greater variation due to Alternative Splicing – different combinations of the exons used to code for the final protein. Alternative splicing creates the potential for multiple protein product from a single gene. For example, some proteins might use the code from all the gene’s exons, but others might use only a few exons, the others having been spliced out, leading to very different proteins with different functions.
Getting back to Intron Retention, this is the situation when an intron escapes being spliced out, and erroneously remains in the mature mRNA. It can happen due to a mutations at a splice site – sequence of DNA that marks where to splice – and therefore, makes the intron invisible to the Splicosome.
The study, published in the November 2015 journal of Nature Genetics, by Hyunchul Jung at the National Research Center in Gyeonggi-do, South Korea, describes the analysis of DNA samples from 1812 patients, with a variety of different types of cancer, including breast, colon, lung, kidney, ovarian, and uterine. After computational analysis of the tumor DNA, Jung found that 31.6% had mutations disrupting normal splicing, with one of the most common type of splicing error being Intron Retention.
Jung also showed that the mutation disrupting the splice site doesn’t need to be one that would change the amino acid sequence of the future protein – a so called Synonymous Mutation- so might be easily overlooked as being disease causing. In many cases, the intron contains a warning signal for the cell to destroy the mRNA before making it into protein, so that gene never gets expressed. In a significant number of the cancers studied, Intron Retention was found to have occurred more frequently in Tumor Suppressor Genes (TSG). TSGs are like the brakes on the cell cycle, and Intron Retention is like having bad brakes, so the cell cycle speeds out of control, leading to a malignant cell that divides uncontrollably. As a suggestion for future research the paper states, “…intronic splice sites should be carefully considered for their potential as disease-causing variants, regardless of whether an amino acid change occurs.” Knowledge of this mechanism of gene disruption may lead to a much better understanding of the causes of certain cancers and other diseases.
Reference Journal:
- Intron retention is a widespread mechanism of tumor-suppressor inactivation. Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. Nat Genet. 2015 Nov;47(11):1242-8. doi: 10.1038/ng.3414. Epub 2015 Oct 5. PMID: 26437032



